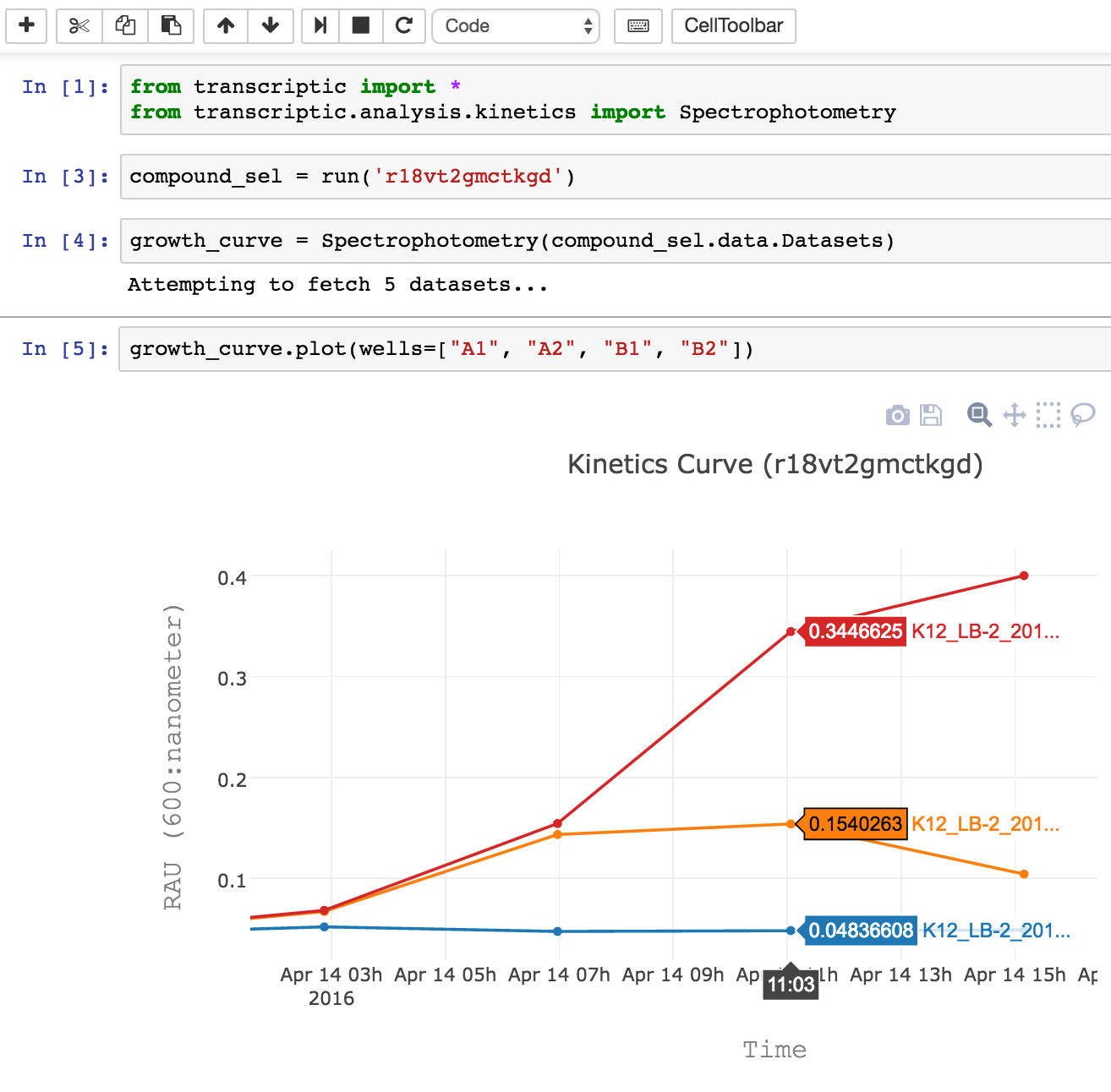
Yang Choo
Hi, I'm a full stack engineer currently obsessed with solving the reproducibility crisis in biology with robotics and machine learning.
I'm leading a team at automated cloud laboratory, working with roboticists, scientists and engineers.
Previously, I studied Chemical Engineering, Biomedical Engineering, and Computational Biology at Carnegie Mellon. At CMU, I was heavily involved in academic research and founded and led multiple organizations to success.